Genetics
Explore inheritance patterns, genetic mapping, and modern genomics. Cross organisms in Punnett squares, perform three-point crosses, and simulate genetic drift in finite populations. These interactive simulations cover classical Mendelian genetics, population genetics, quantitative trait loci mapping, genome-wide association studies, and cutting-edge sequencing technologies.
Classical & Mendelian Genetics
Build Punnett squares and explore dominance, recessiveness, and epistasis. Trace inheritance through pedigrees and perform genetic linkage crosses to map gene order.
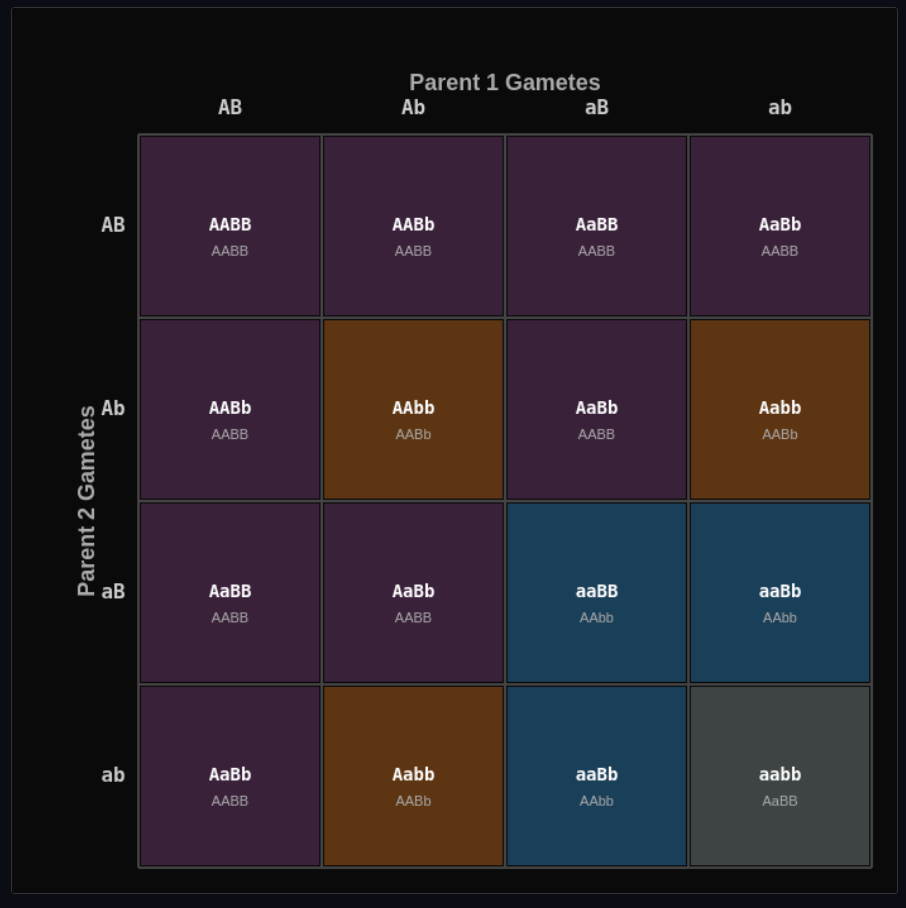
Mendelian Genetics Sandbox
Cross organisms and build Punnett squares—explore dominance, recessiveness, and epistatic interactions
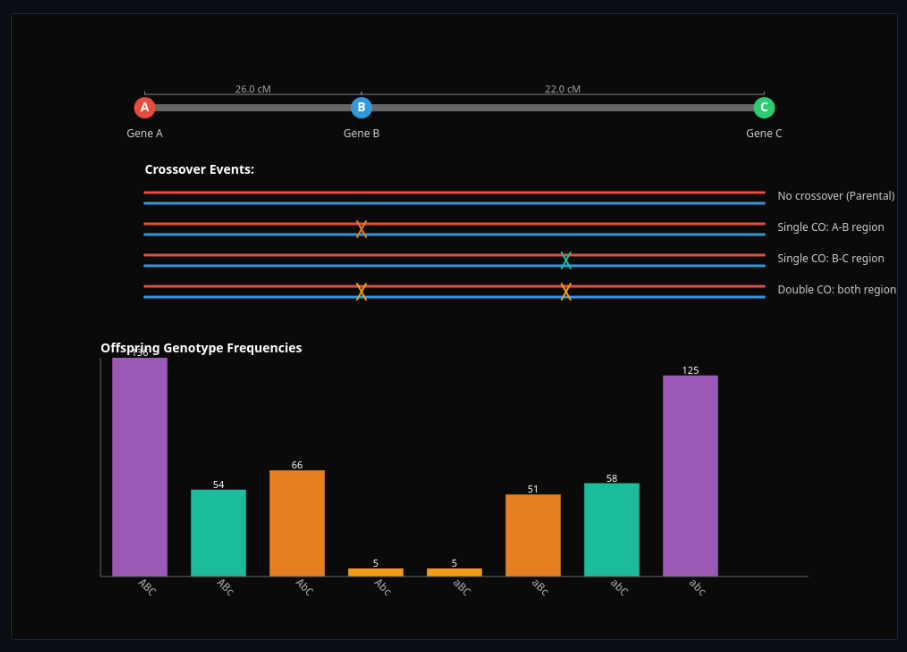
Genetic Linkage & Recombination
Perform three-point crosses to map gene order—account for crossover interference in recombination
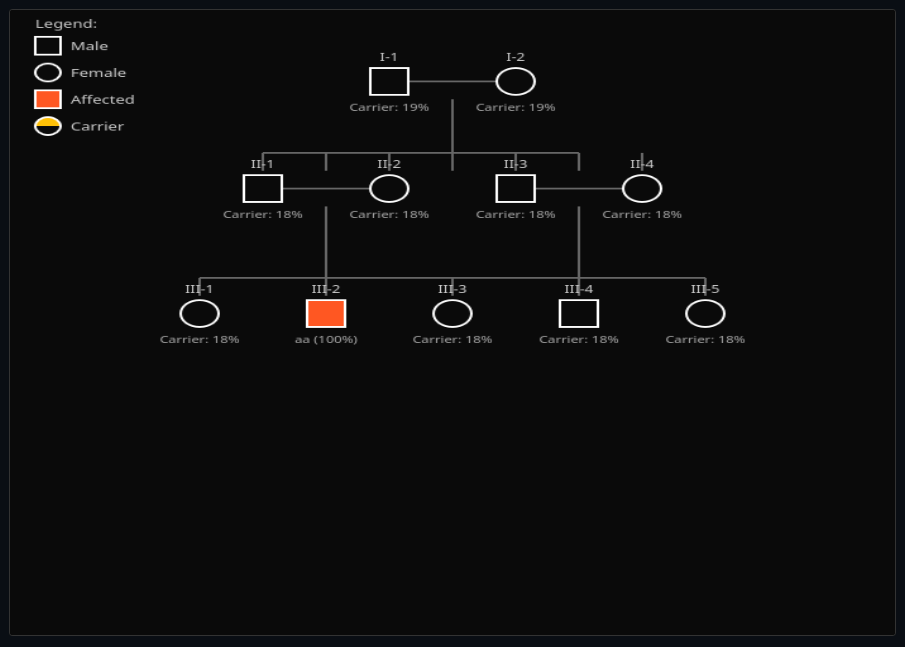
Pedigree Inheritance Analysis
Trace inheritance through family trees—calculate carrier probabilities for recessive diseases
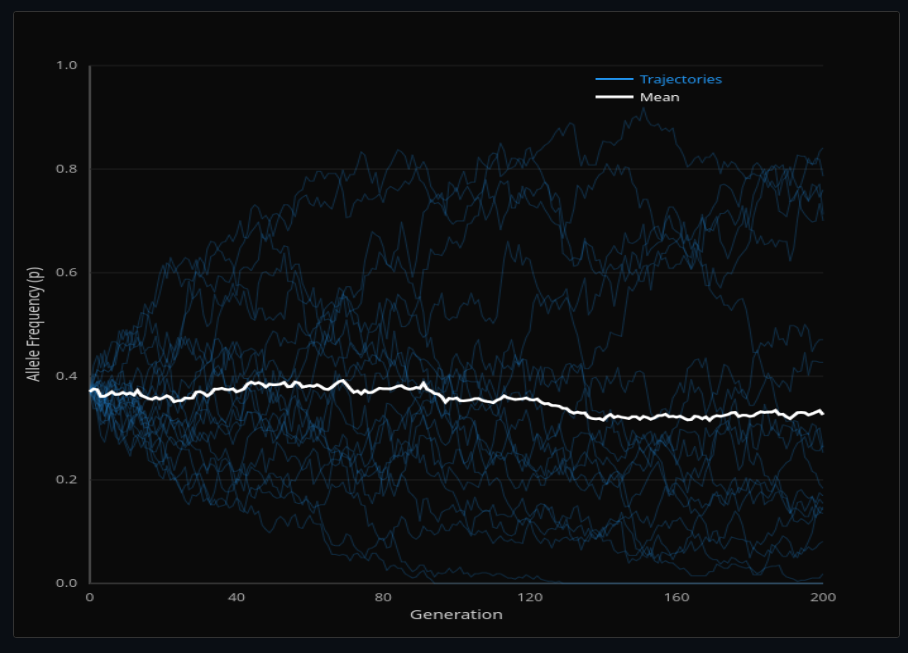
Population Genetics
Simulate genetic drift with the Wright-Fisher model and compute identity by descent coefficients. Watch alleles randomly fix or go extinct in finite populations.
Genetic Mapping & QTL Analysis
Design F2 and backcross experiments to build genetic maps. Perform interval mapping to identify quantitative trait loci and run genome-wide association studies.
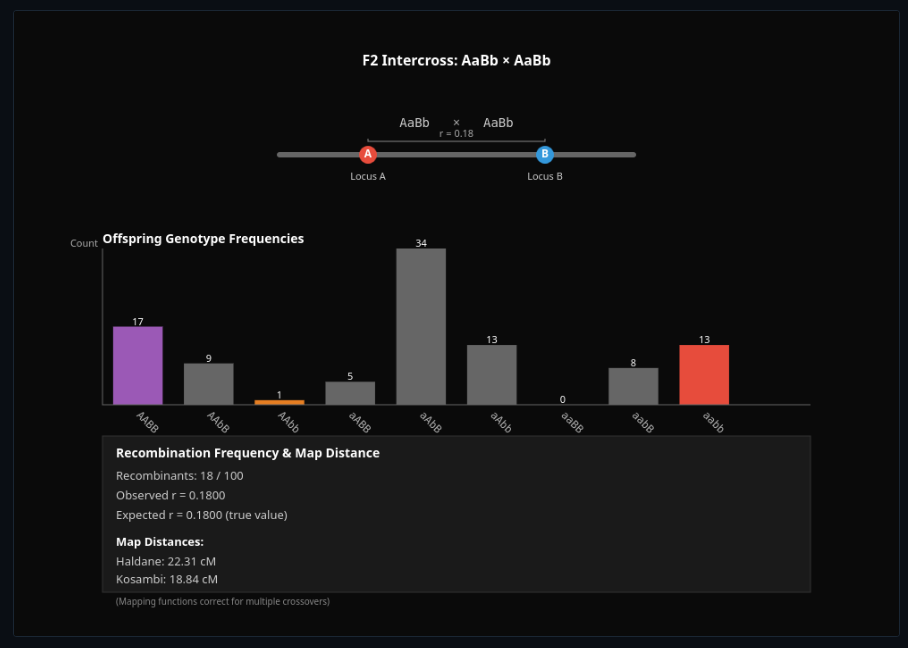
Genetic Linkage Map Builder
Design F2 or backcross experiments to estimate recombination frequency—build genetic maps from segregation data
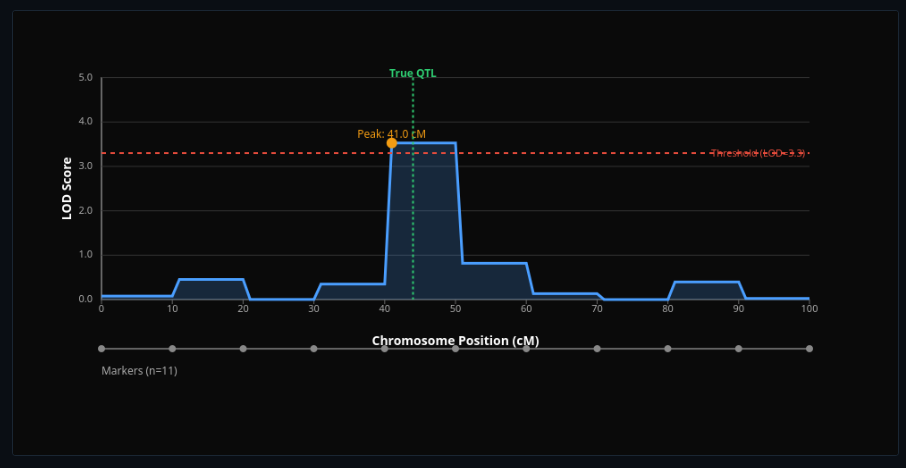
QTL Interval Mapping
Scan the genome for quantitative trait loci—plot LOD scores and identify QTL intervals
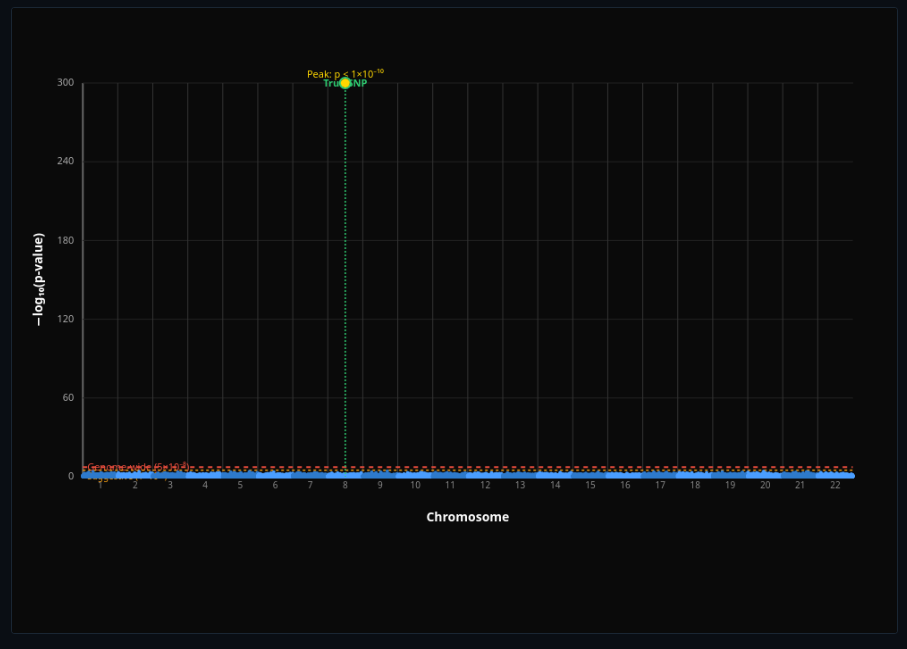
Genome-Wide Association Study
Run genome-wide association tests and plot Manhattan plots—adjust sample size to see power increase
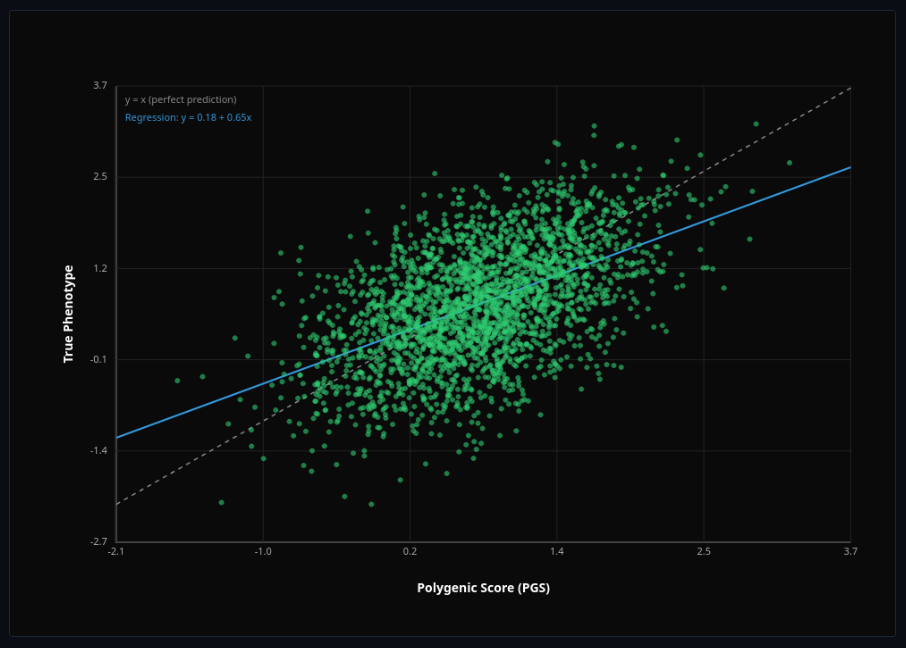
Polygenic Score
Prediction accuracy and portability
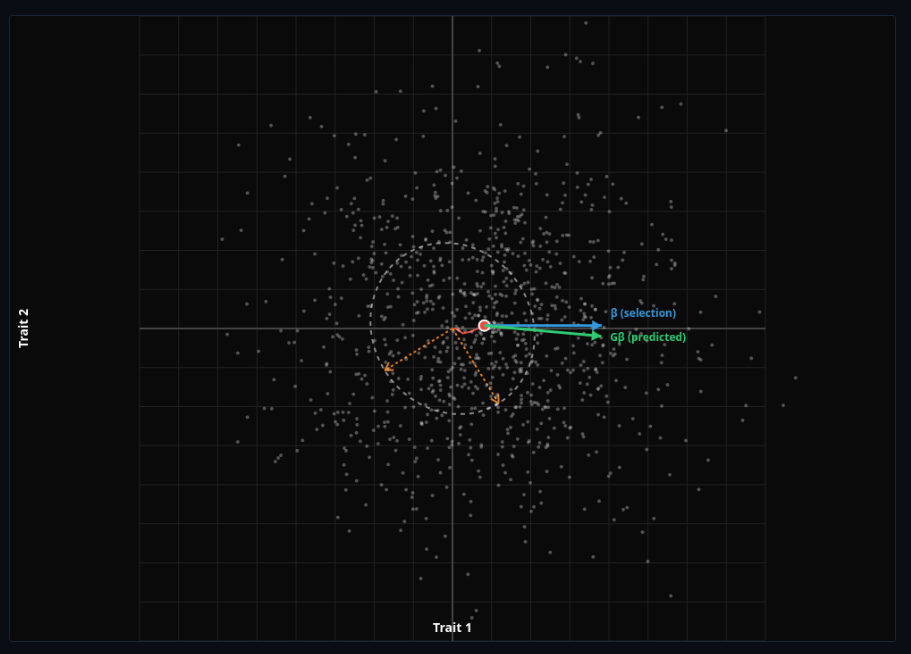
G-Matrix Evolution
Multivariate quantitative genetics
Molecular Genetics & Gene Expression
Analyze codon usage bias and watch ribosomes traffic along mRNA. Explore alternative splicing patterns, transcription factor combinatorics, and chromatin state dynamics.
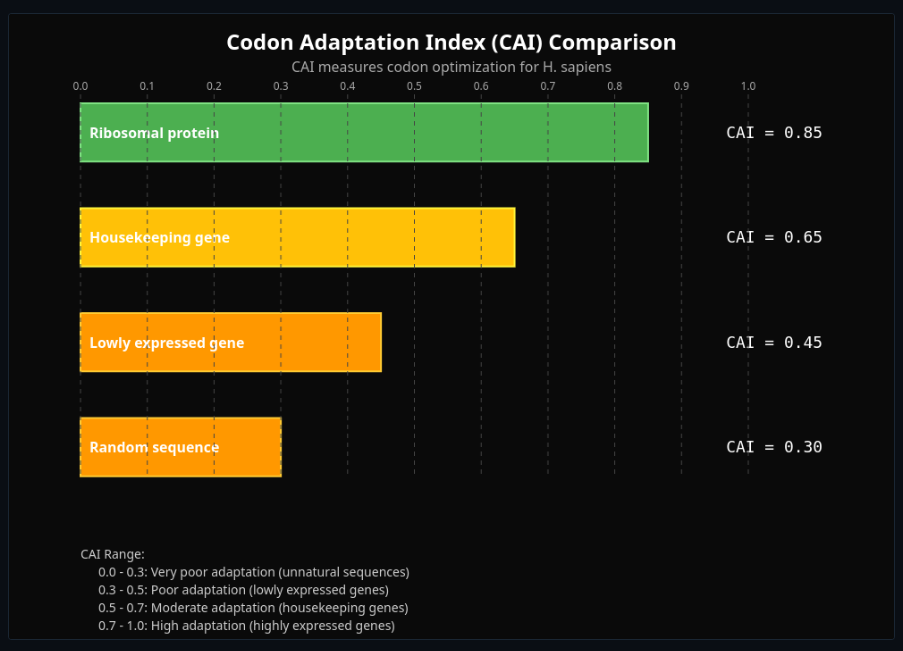
Codon Usage Bias & tRNA Adaptation
Analyze codon usage bias and tRNA adaptation—compute CAI and see translation efficiency
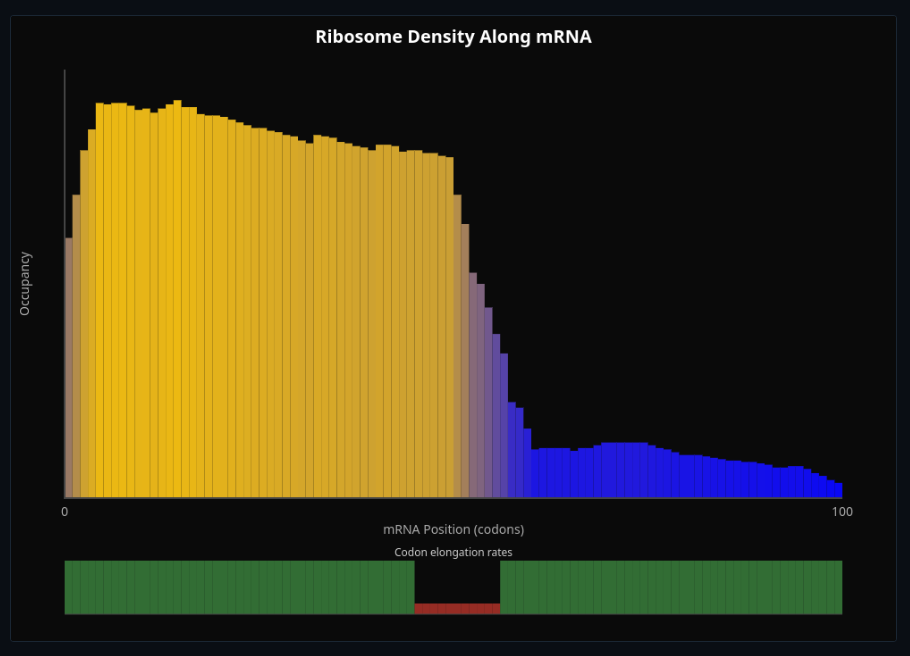
Ribosome Traffic & Translation
Watch ribosomes traffic along mRNA—see polysome formation and translation kinetics
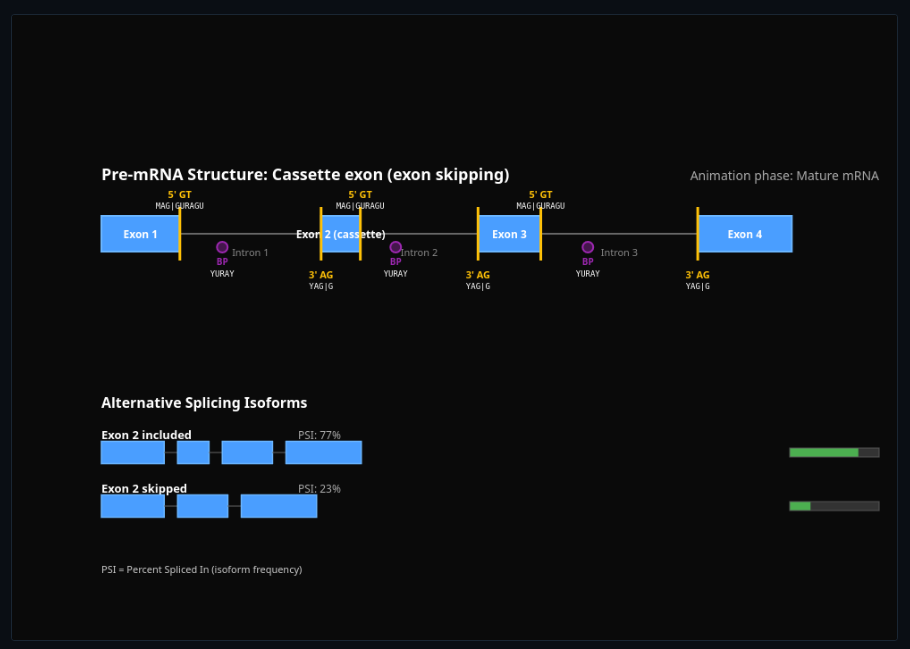
Alternative Splicing Simulator
Splice exons together with different patterns—explore constitutive and alternative splicing mechanisms
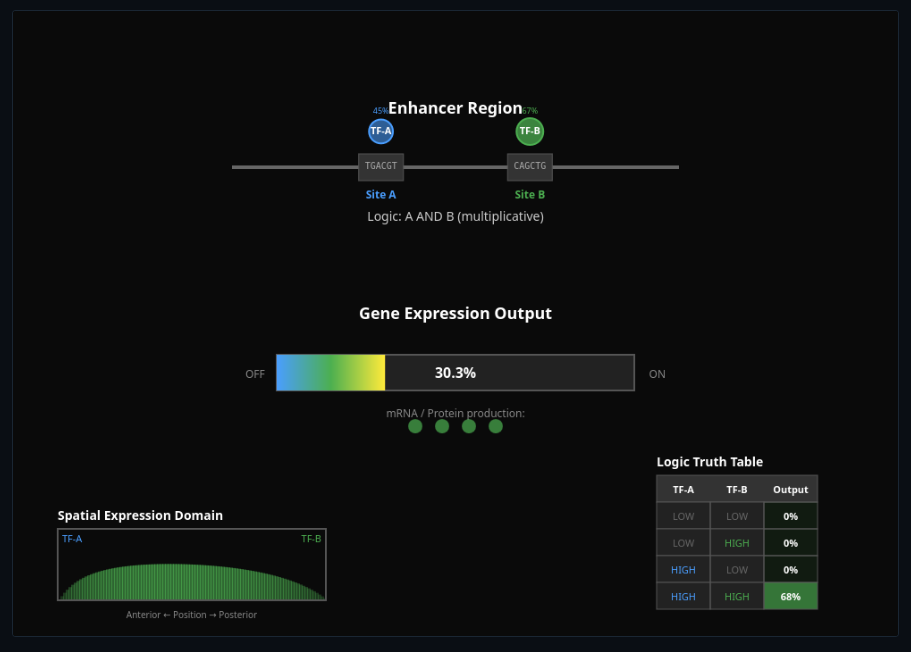
Enhancer Logic & TF Combinatorics
Combine transcription factor binding sites—see how combinatorial TF logic controls gene expression
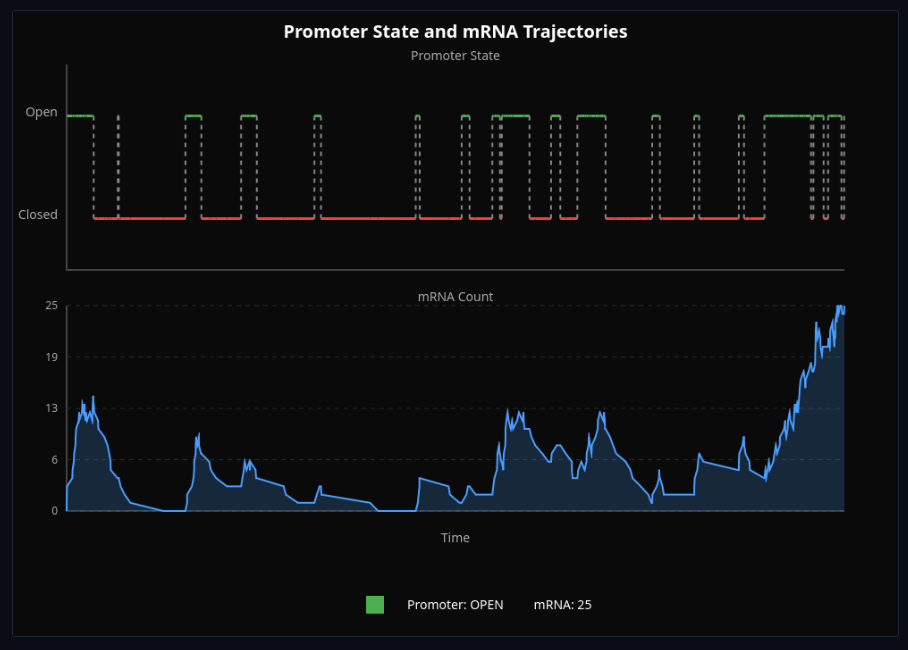
Chromatin State & Transcriptional Bursting
Toggle chromatin between open and closed states—watch transcriptional bursting from stochastic promoter switching
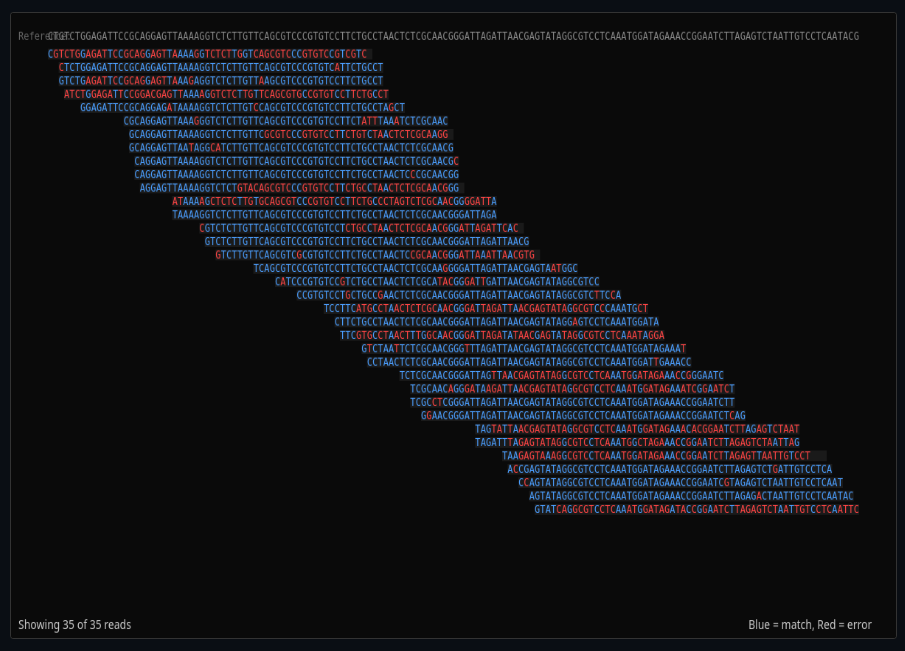
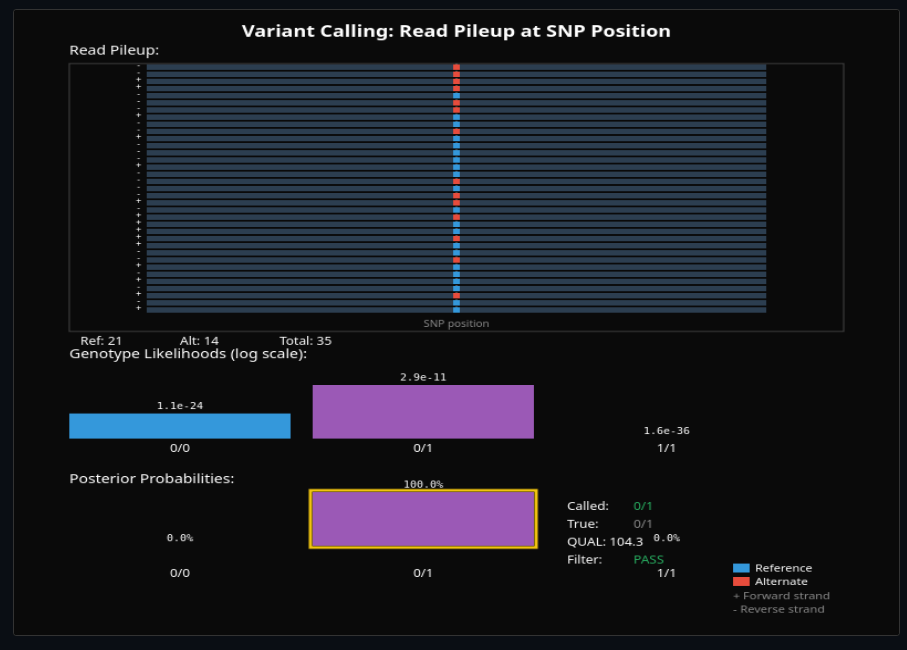
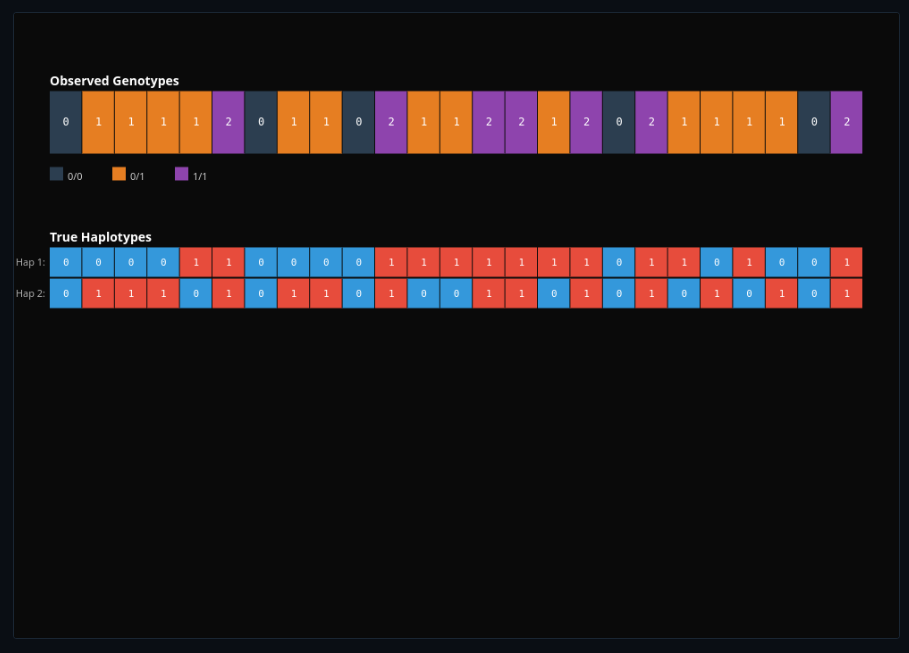
Genomics & Sequencing Technologies
Simulate DNA sequencing reads with errors and quality scores. Call variants, phase haplotypes, and assemble genomes from de Bruijn graphs.
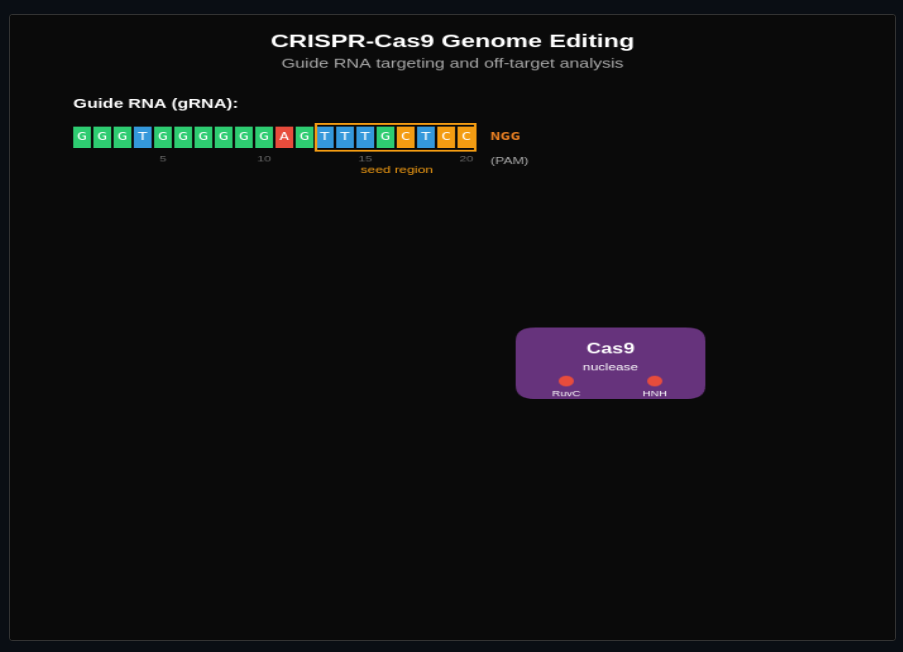
Genome Engineering
Design CRISPR guide RNAs and explore on-target vs off-target editing. Model NHEJ and HDR repair outcomes from double-strand breaks.